Case Study 2.1: How do UberEats and Google Maps speedup geo queries hashing in RAM?
How do UberEats and Google Maps speedup geo queries using hashing in RAM?
Goal: Learn how to build indices on multidimensional data
In food delivery and mapping applications, it’s important to optimize the travel times and cost. Consider how to group "nearby" orders together and assign them to the same delivery driver.
For example, let’s say we have a list of orders with the following locations in latitude, longitude:
orders = [(37.788022, -122.399797), (37.788122, -122.399797), (37.788022, -122.398897)]
Geo data often involves searching for points that are near each other in two dimensions: latitude and longitude. To accomplish this, we need to extend our hashing and sorting techniques beyond the one-dimensional search key, such as a song ID, to account for nearness.
Geo Hashing
Geo hashing is a method for encoding a geographic location into a short string of letters and digits.
-
Grid Division: We divide the earth into a grid of squares, each identified by a unique geo hash.
-
Recursive Sub-division: We divide each square into smaller squares, repeating the process until we reach our desired precision (geo hash length).
Example: For (37.788022, -122.399797) with a length of 5 characters (~2.8 km precision), the resulting geo hash is "9q8yy". This hash can be used to quickly find nearby locations with small variations of the string.
Hexagon-based Hashing (Uber H3)
Hexagon-based hashing using H3 cells is a recent and more powerful method (see H3 vs S2 comparison). H3 is a hierarchical grid system that uses hexagons to divide the earth's surface into cells of various resolutions.
You can experiment with these techniques in this Hashing Colab.
Why Hexagons?
Unlike squares, hexagons have the same distance between a hex's center and the centers of all its neighbors. This makes them ideal for radius-based searches and smoothing.
Uber H3 in Action:
At zoom level 8, many close locations map to the same cell ID. At zoom level 12, locations that are very close (like buildings on a campus) might differ only in the last few characters of their cell IDs.
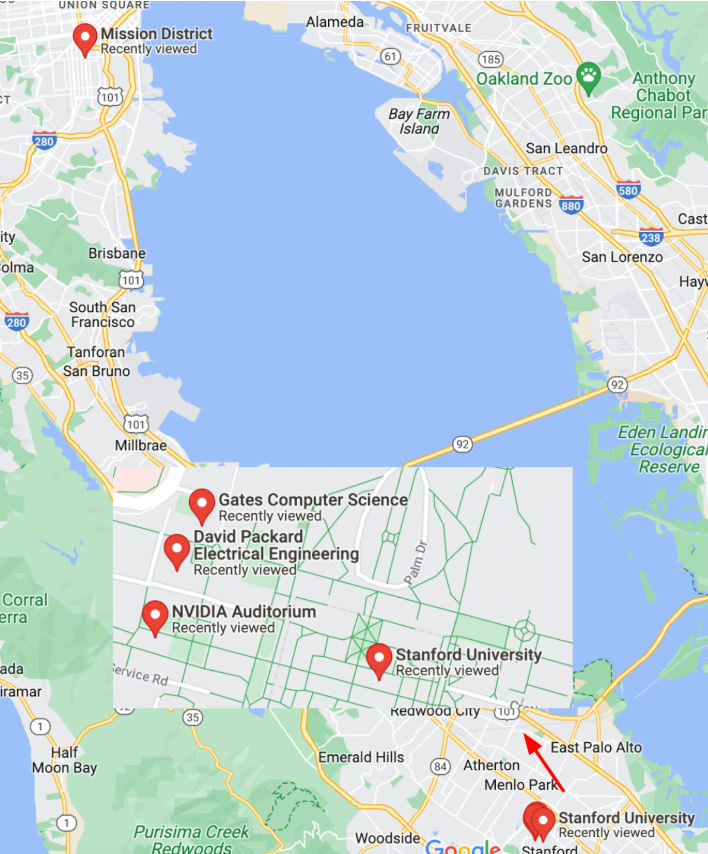
This code snippet shows how to use the h3.geo_to_h3 function to convert latitude and longitude coordinates into hexagonal cell IDs for a given zoom level. The example includes the coordinates of four locations: SF Mission, and three buildings on Stanford campus (Nvidia Auditorium, Packard and Gates buildings).
import h3
# lat/lngs from SF mission, Stanford NVidia, Packard, Gates Buildings
locations = [(37.788022, -122.399797), (37.428226, -122.174722),
(37.429749, -122.1735490), (37.429761, -122.173290)]
for zoom in [8, 12]: # zoomlevels 8 and 12 (imagine the +/- in map zoom)
for l in locations:
cell_id = h3.geo_to_h3(l[0], l[1], zoom)
print(f"@zoom[{zoom}]: {cell_id}")
Output:
@zoom[8]: 88283082a1fffff
@zoom[8]: 8828347417fffff
@zoom[8]: 8828347417fffff
@zoom[8]: 8828347417fffff
@zoom[12]: 8c283082a06bbff
@zoom[12]: 8c28347416257ff
@zoom[12]: 8c2834741602bff
@zoom[12]: 8c28347416021ff
At zoom level 8, all three Stanford locations map to the same cell ID. At zoom level 12, Packard and Gates, which are close to each other, differ only in the last three characters of their cell IDs. Similarly, NVidia differs from the other Stanford locations only in the last five characters of its cell ID, while SF Mission, which is further away, has a unique cell ID.
Uber Eats (and each geo app) picks one or more zoom levels for hashing, based on the use case and the required precision.
Extending Hashing to More Domains
1. Perceptual Image Hashing (PhotoDNA)
PhotoDNA (from Microsoft) helps detect known illegal content. It creates a robust hash that identifies an image even if it has been resized, compressed, or had color changes. Similar open-source techniques include imagehash.
2. DNA Sequence Hashing
Hashing DNA sequences involves Window Hashing (breaking sequences into overlapping substrings of length k, called k-mers). These k-mers are then hashed (e.g., using SHA-256) to create unique values for fast partitioning and indexing of genetic data.
Takeaway: By mapping complex multidimensional data (Geo, Images, DNA) into hash values, we can efficiently index and retrieve data while minimizing IO operations.